- Get link
- X
- Other Apps
I wake-up every day with an insatiable desire to propel the idea of Individualized Signalling Biology which is poised to facilitate Personalized Medicine in Immuno-Oncology. Individualized Signalling Biology is based on the idea that coding region single nucleotide polymorphism (cSNP) variants of cell surface receptors drive non-canonical signalling pathways that are unique to an individual's genotype.
An idea that got its seeds planted while working with the Lasker Award winner Axel Ullrich, Director of the Department of Molecular Biology at the Max-Planck-Institute of Biochemistry (MPIB). After Tony Hunter discovered tyrosine phosphorylation in 1979, it remained unclear on how this rare event was triggered inside living cells. Few years later, Axel Ullrich provided the first concrete evidence evidence for the existence of transmembrane proteins containing an extracellular segment to detect signals outside the cell and an intracellular segment with tyrosine kinase activity to transduce signals inside the cells. His seminal work laid the foundation of signal transduction by means of receptor tyrosine kinases (RTK) across cell membranes and also led to the development of the first molecularly targeted drug and first personalized medicine (Trastuzumab for patients with ERBB2 over-expressing tumors). He identified many genetic mutations in several RTKs some of which were found to be driving the tumor growth in animal models. One among them was the germline variant FGFR4 p.G388R, which is associated with almost all kinds of cancer types. I joined his team in 2010. Soon thereafter I detected the existence of an individual genotype (rs351855-A)-specific tyrosine phosphorylation signalling pathway in human cells. I continued working with Axel Ullrich even after his retirement in 2016.
Currently, I am heading a research group hosted by the University Medical Center in Göttingen, Germany. A
typical work day in my research group begins with an aromatic cup of
freshly brewed coffee ☕ and raging thoughts on new hypotheses for
experimentation 👨🏽🔬 🔬. Luckily, all my students are quite
articulate and never shy from asking provocative questions that we love
to answer every week. I tend to think along the lines of Individualized Signalling Biology and use every opportunity I get to popularize this concept within the campus; by collaborating with clinicians and basic science research groups and by designing new elective lectures on this topic. Additionally, I am also mobilizing graduates, postgraduates, doctorates, post-doctorates and independent investigators to collaborate with me and participate in research projects related to Individualized Signalling Biology. If you are interested in a collaboration, or your students in a joint-lab internship or joint-lab rotation in my research group, please don't hesitate to send me an Email. I will be happy to discuss potential opportunities for joining forces.
Individualized (precision) signalling biology is a fascinating new concept which is currently at its infancy. I have been pursuing this unique and very challenging area of research for the past 10 years.
A simple immunoblot analysis of human primary breast epithelial cells to assess STAT3 signalling pathway reveals significant differences between two individuals. Such differences in other signalling pathways are also expected when working with same cell types derived from unrelated human individuals.

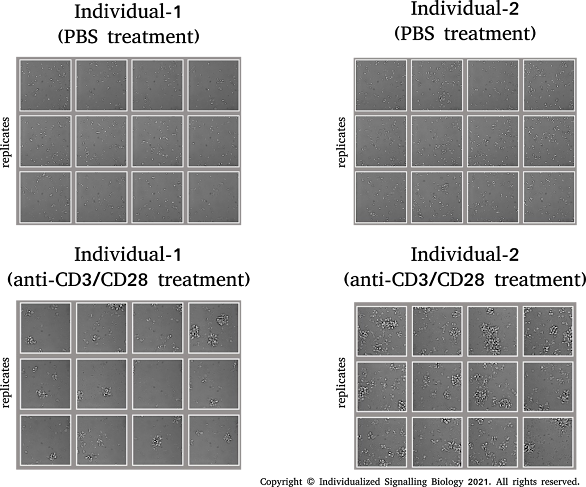
It is not just the activation of signalling molecules, differences in signalling outcomes are also often observed in primary cells derived from different human individuals. Yes, that is true. When examining the same signal transduction pathways, in the same cell types which are isolated, prepared and handled in exactly the same way, one can encounter differences in the signalling outcomes. For instance, below I show an image depicting the proliferation capacities of CD8 T cells which were isolated from the blood donated by two gender & age-matched human individuals. An experiment that was done at the MPIB in 2017 with the scientific co-operation of the Institute Doctor (Betriebsarzt: Dr. Reichert, J).
Purified CD8 T cells from two individuals were treated with identical amounts of anti-CD3 and anti-CD28 agonistic monoclonal antibodies and (mAbs). These mAbs specifically stimulate T cell receptor (TCR) complex to induce signalling across the T cell membranes by increasing the tyrosine phosphorylation of all key molecules involved in controlling T cell proliferation. PBS treatment serves as vehicle control. The outcome of TCR stimulation can be measured after 3 days by looking for cellular proliferation.
What you can see here is that triggering the same cell surface receptor i.e., TCR, in same cell type i.e., CD8 T cells, but derived from different individuals did generate different TCR signalling outcomes. Individual-2 had better proliferative response as compared to that of Individual-1. It is very uncommon to encounter such differences if the same experiment was to be conducted using CD8 T cells derived from gender & age-matched animal models such as mouse or rat. One can appreciate that there is an important need to figure out the molecular factors governing such individual-individual differences at the signalling level. These differences are not trivial and one must take into account when extrapolating findings from basic science research to human cells.
Clinicians and trained medical professionals who work with patient materials or conduct clinical trials are very well aware of this problem. "Standard Deviation", "Heterogeneity" and "Outliers" are the buzzwords in the world of clinical and translation-oriented research.
Yet, "One Size Fits All" approach was the generally accepted paradigm in human biology for a very long time. I want to challenge this paradigm. Thanks to the wealth of genetic data generated by various individual genome sequencing projects, that this vision now seems very plausible. There are many programs that are uncovering thousands of new missense cSNP variants almost on a regular basis. Some well known programs include the All of Us Research Program by the National Institute of Health; the Personal Genome Project by the Harvard Medical School and the Human Genome Diversity Project by the Stanford University.
Have you ever thought about why then biologists still use “model organism?” Of course, conducting basic research on model organisms helps researchers gather a general perspective of all evolutionarily conserved aspects of the cellular and molecular biology of the human body. However, the real reason is convenience and practicality. Model organisms have characteristics that allow them to be easily maintained, reproduced, manipulated and studied in a well-controlled laboratory conditions. The most common model organisms used for basic science research are fruit-fly, worm, zebra-fish, mouse and rat. An important point not to forget here, that model organisms used for research are generally inbred animal species and therefore genetically homogeneous. Now compare that with the biology of humans, who are genetically heterogeneous and out-bred species, meaning every human individual is different from every other individual, estimated to be at about ~4 - 5 million differences (genetic variation). This means an individual is different from another at about 4 to 5 millions places in their genomes. Therefore, certain aspects of signalling biology applicable to human individuals can never be uncovered studying only model organisms.
Individualized approach to signalling properties of cell membrane proteins is therefore an innovative research concept that is poised to fill this gap in our understanding of individualized signalling biology. My research focuses on cell membrane proteins as they are responsible for almost all diseases and thus are frequently targeted for therapeutic interventions. I want to understand why therapies do not work for some individuals and why they cause more side-effects to some of us. To this end, I take interdisciplinary approaches to elucidate human receptor variant-specific signalling pathways. Subsequently, this should facilitate the realization of the full potential of individualized or personalized medicine.
I am sure many experts can agree upon that.
Consider this. Our body is made up of ~3.7 trillion cells and thousands of proteins, nobody knows the exact number but a modest estimate is about ~25,000. If humans are all made up of same number of cell types and same number of proteins why do we see differences among individuals at the molecular, cellular, organisms and population levels? The differences arise due to genetic heterogeneity which generates differences in quantities and qualities of protein sequences and thereby their functional properties within each cells. This means, although we are all made up of same number of proteins, they are not identical in each individuals. Every individual is made up of different versions (sequence variants) of same proteins and often found in differing amounts inside our cells.
Ultimately it is these protein variants and their unique variant-specific functional properties that dictates our look, behavior, susceptible to diseases, response to medications or for some the super human skills.
In other words, protein variants determine our uniqueness in fighting diseases or responding to drug induced toxicities or adverse effects.
Therefore,
if we are serious about
realizing the goal of individualized medicine, it is important to investigate the biology of human protein variants and uncover
the protein variant-specific signalling properties.
So next time you come across a paper or a project that deals with membrane proteins, just think about how many individual-specific variants with unique structural & functional properties might exist out there in the general population. Ask yourself the question, if the time is right for individualized approach to signalling biology?
If you like what you read, please consider clicking that "Share" / "Follow" link or post your comments below.
Copyright © Individualized Signalling Biology 2021. All rights reserved.
Cancer
Cell Membrane Proteins
Immuno-oncology
Immunology
Individualized Biology
Individualized Medicine
Precision Medicine
Receptor Variants
- Get link
- X
- Other Apps
Comments
Post a Comment